Natural Genetic Modification and Hazards of GMOs
A culture of denial over the horizontal spread of genetically modified nucleic acids prevails in the face of direct evidence that it has occurred widely when appropriate methods and molecular probes are used for detection Dr. Mae-Wan Ho
This article has been sent to Dr Kaare Nielsen in his capacity as a member of the European Food Safety Authority GMO Panel and he is given the right to reply
The first genetically modified (GM) crop was commercially approved and released into the environment 20 years ago. From the beginning, some of us have been warning repeatedly of hidden dangers from the unintended horizontal transfer of GM DNA (transgenes). A comprehensive review [1] Gene Technology and Gene Ecology of Infectious Diseases, ISIS scientific publication) and successive updates were submitted to the World Health Organization (WHO) and regulatory agencies in the US, UK and European Union (see [2] Ban GMOs Now, ISIS Report); all to no avail.
The position taken by regulators and their scientific advisors today is perhaps best represented in a recent publication [3] with lead author Kaare Nielson at Norwegian University of Science and Technology, who both advises Genøk-Centre for Biosafety and serves as member of the EFSA (European Food Safety Authority) GMO panel.
The paper, entitled “Detecting rare gene transfer events in bacterial populations”, recognizes that horizontal gene transfer is part of the risk assessment for GMOs, and that the large-scale cultivation of GM-plants on more than 170 m ha worldwide results in “multitudinous opportunities for bacterial exposure to recombinant DNA and therefore opportunities for unintended horizontal dissemination of transgenes.” It admits that horizontal gene transfer has indeed been demonstrated in the laboratory. “But in natural settings, negative or inconclusive evidence has been reported from most sampling-based studies of agricultural soils, runoff water and gastrointestinal tract contents.”
It tells us that horizontal gene transfer research “suffers from significant methodological limitations, model uncertainty and knowledge gaps.” In particular, on account of the “low mechanistic probability of horizontal transfer...in complex environments”, it would take “months, years, or even longer for the few initially transformed cells to divide and numerically out-compete non-transformed members of the population” for them to be “detectable.” The rest of the paper mentions a mathematical model based on those very assumptions, the most important being the very low probability of horizontal transfer; which has been contradicted by empirical evidence, most decisively from a study in China reported in 2012 [4].
I have reviewed the positive and circumstantial evidence for horizontal transfer of GM nucleic acids thoroughly in [2]. The present report updates on important new developments.
Nielsen is also a lead investigator of a new study [5] showing that even short and damaged DNA, ubiquitous in most environments, is readily taken up by many bacteria and incorporated into the bacterial genome. The results not only contradict his earlier claim that horizontal transfer of transgene DNA has a “low mechanistic probability”, but also previous work claiming that only much longer sequences are effectively transferred. The new study shows that damaged and degraded sequences as short as 20 bp could be taken up and incorporated into the bacterial genome, including DNA from a 43 000 y old woolly mammoth bone. The significance of the research is highlighted [5] (p. 1): “Our findings suggest that natural genetic exchange of DNA from dead and even extinct organisms to contemporary bacteria can take place over hundreds to thousands of years. Hence damaged and degraded DNA may be a previous unrecognized driver of bacterial evolution with implications for evolutionary theory.”
But no mention is made of the huge amounts of transgenic DNA released into the environment from GM crops and industrial ‘contained’ uses of GMOs, which most certainly could be taken up and incorporated into bacterial genomes by similar mechanisms.
Nielsen and colleagues point out that natural transformation with short DNA fragments are likely to involve base substitutions that result in modification or loss of gene function, rather than acquisition and integration of entire genes that happens in transformation with long DNA fragments. (They fail to point out that promoter/enhancer sequence of genes and sequences encoding the plethora of newly discovered regulatory RNAs [6] can be very short indeed, and when taken up and incorporated into the bacterial genomes, can have sweeping effects on gene expression and biological functions, including those involved in causing diseases to plants, animals and humans.)
More importantly, short DNA molecules do not encounter the same barriers to natural transformation as longer DNA fragments such as conflicting gene order, function and DNA similarity. Although requirements for DNA similarity are still present, short similar DNA stretches are more likely to be found in a broader range of species, and the probability of random sequence similarity increases as fragment size decreases.
The transformation frequencies of short DNA sequences are relatively low compared with long sequences; but, as the authors point out, short DNA sequences are much more plentiful in the environment than long sequences, because the majority of DNA sequences will be in various states of degradation.
Nielsen and colleagues highlight the fact that two adjacent nucleotide changes - which they find to be more likely than single nucleotide changes - can result in markedly increased antibiotic resistance, and “[b]iological waste disposal and decontamination practices, e.g., in hospitals where antibiotic-resistant infections are common, are focused on controlling organisms rather than free DNA molecules, and usually results in only partial DNA fragmentation.” In that context, the authors could have included laboratories and contained facilities creating and growing GM microorganisms from which large amounts of nucleic acids are routinely discharged into the environment as ‘tolerated releases’ [7] (SLIPPING THROUGH THE REGULATORY NET: ‘Naked’ and ‘free’ nucleic acids, I-SIS/TWN report).
The authors constantly allude to “natural transformation” to emphasize the increasingly accepted notion that horizontal gene transfer is part of the amazing feats of ‘natural genetic engineering’ or ‘natural genetic modification’ that organisms and cells carry out in order to survive (see [8] Evolution by Natural Genetic Engineering, SiS 63). However, there is nothing “natural” about GM nucleic acids, which contain numerous synthetic parts and novel combinations, and designed to overcome and override natural genetic modification processes. Nevertheless, GM nucleic acids can exploit natural transformation processes to wreak potential havoc on health and the environment [2, 9]. There is already evidence that short, degraded DNA is readily taken up by human cells and integrated into the genome; that nucleic acids are actively secreted by cells into the circulatory system, and nucleic acids in food can enter the blood stream and influence gene expression in cells of the body (see [10] Nucleic Acid Invaders from Food Confirmed, SiS 63).
Nielson and colleagues [3, 5], while not denying that horizontal transfer of GM nucleic acids can take place, is giving the wrong impression that it has a very low probability, hence lulling the public into a false sense of security, thus giving licence to continued release of GMOs into the environment. Sadly, they have been basing their arguments on obsolete assumptions concerning the transformability of bacteria and the probability of horizontal gene transfer, as well as outdated technologies for detecting horizontal gene transfer which were highly insensitive [3]. They have been ignoring new findings, including their own, and the fact that the technologies have advanced by leaps and bounds in recent years. It is now possible to detect horizontal gene transfer directly without culturing the bacteria (see [2]); this is important as the overwhelming majority of bacteria in the environment cannot be cultured in the laboratory, and consequently, direct detection is revealing much higher frequencies of horizontal gene transfer. In addition, PCRs (polymerase chain reactions) for detecting specific sequences and nucleic acid sequencing methods have improved to such an extent that they can even be done on single cells [11]. Finally to detect GM DNA, it is necessary to use the appropriate molecular probes (primers) and analysis based on the relevant bioinformatics databases. Using such methods, scientists in China have indeed detected specifically the antibiotic resistance marker gene in commercial plasmids that is turning up in all of China’s rivers [4] (see below), even though the country has not been commercially growing GMOs to any great extent.
The assumption that only some bacteria are naturally transformable is questionable, another indication that horizontal gene transfer via uptake of nucleic acids is much more widespread. Researchers led by Dongchang Sun at Zhejian Aademy of Agricultural Sciences Hongzhou China, have discovered that ‘non-transformable’ species of bacteria such as the common laboratory bacterium Escherichia coli is readily transformable by plasmids (small independently replicated genetic structures, usually circular, and much used in genetic manipulation) [12]. Gene transfer through plasmid is one of the major routes for spreading antibiotic resistance in bacteria. Outbreaks of life-threatening superbugs such as NDM-1 bacteria and enterohaemorrhagic Escherichia coli (EHEC) are often due to the transfer of antibiotic resistance genes in plasmids. Although plasmid transfer by conjugation (a bacterial mating process) was discovered in E. coli long ago, the species was traditionally considered not naturally transformable and needs special treatments such as electric shock or Ca2+ stimulation and heat shock. Yet, a complete set of competence gene homologs for DNA-uptake machinery is found in the genome of E. coli. Moreover, transcription of some competence genes is inducible by a competence regulator homolog Sxy. The team has previously shown, as others have, that E. coli is able to acquire naked plasmid DNA on agar plates at 37 °C without any special treatment. Spontaneous plasmid transformation of E. coli is independent of the DNA uptake machinery for single-stranded DNA entry. Instead DNA is taken up rapidly within 2 minutes of exposure, and most of the transforming DNA will have entered E. coli cells within 10 minutes.
At the end of 2012, Li Jun Wen, Jin Min and colleagues at Sichuan University in China reported finding genetically engineered plasmids containing an antibiotic resistance gene in all 6 of China’s rivers [4]. We have reported extensively on it [13] GM Antibiotic Resistance in China's Rivers (SiS 57), including a list of GM crops already commercialized, field tested, or imported that contain the particular ampicillin resistant gene blá.
What is so significant about the work is that the researchers set out specifically to investigate if the genetically engineered antibiotic resistance gene is present in the environment, using the appropriate molecular probes and tools that are readily available. And it is the first ever such study in the world. Consequently, claims such as that made by Nielson and colleagues [3] and other regulatory agencies and promoters of GMOs that horizontal transfer of transgenes does not take place, or only very rarely takes place in nature is fully contradicted. They simply have not been looking for it; it is a case of “don’t look, don’t find.”
The Sichuan University research team wrote ([4] p. 13448): “While antibiotic-selectable synthetic plasmid vectors have proved invaluable tools of genetic engineering, this class of artificial recombinant DNA sequences with high expression of antibiotic resistance genes presents an unknown risk beyond the laboratory setting.”
Using PCR (polymerase chain reaction) and real-time quantitative PCR with a combination of primers designed to detect the synthetic plasmids containing the antibiotic resistance gene, they detected various levels of blá in all six rivers sampled, with the highest levels in the Pearl and Haihe rivers (Figure 1).
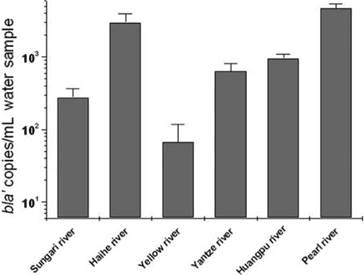
Figure 1 Genetically engineered plasmid ampicillin resistance gene in China’s rivers
The environmental plasmids in the river samples were used to transform E. coli cells to create a ‘plasmid metagenome’ library of 205 ampicillin-resistant strains. Of these, 27.3 % were positive for blá on PCR analysis. Sequencing confirmed the synthetic plasmid vector sources. In addition, tests for antibiotic resistance confirmed the ampicillin-resistant functions of the environment plasmids. The resistance spectrum of transformants from the Pearl and Haihe rivers, in particular had expanded to the third and fourth-generation of cephalosporin drugs, while that of other transformants mainly involved first and second generation cephalosporins.
The researchers explained why they focussed on the synthetic plasmid antibiotic resistance marker (pp. 13448-9): “Plasmids have been exploited as experimental tools to facilitate rapid and efficient genetic engineering. To improve transforming efficiency and clone yields, synthetic plasmid vectors were developed with series of selectable genetic markers, among which antibiotic resistance sequences proved especially useful. Further manipulation to achieve hyper-transmissibility allowed these synthetic plasmids carrying multiple drug resistance genes, to be readily horizontally spread in the laboratory setting.
“Over the past decade, genetic engineering technology has expanded beyond scientific research into practical industries including biofuel fermentation, agriculture, and environment bioremediation. Consequently, the synthetic plasmid vectors used in industrial applications have a greater chance of uncontrolled discharge into the environment, where they may pose a risk of transferring their antibiotic resistance genes to natural microbes.”
The blá recombinant gene was derived from wild-type b-lactam hydrolase genes, which confer robust drug resistance to pathogens and include the extended spectrum b-lactamases and New Delhi metallo-b-lactamase-1 (NDM-1), but which also have high mutations rates.
Three pairs of universal primers were designed for detecting the blá gene and two pairs of primers for pUC and PBR322 synthetic vectors respectively covering insertion sites for sequence detection of the foreign DNA including the recombinant blá gene.
The PCR products of 6 river samples and 6 transformants in the plasmid metagenomic library were selected for sequence alignment and phylogenetic analysis. Alignments hits most frequently represented artificial or synthetic constructs, including cloning expression, shuttle, gene fusion and gene trap vectors. In addition, screening results for vectors (VecScreen) demonstrated that the segments matched most strongly to the pBR322 vector, with sequence identity up to 100 %. Further, systematic (one-by-one) analysis of the matched regions revealed the presence of a partial blá in all samples and transformants.
The team explained their methodology more fully in the discussion (p. 13452): “The blá gene was developed by recombinant technology as a functional fragment (~861 bp) to support cloning applications. Over the years, this antibiotic selective sequence has been introduced into a series of popular cloning vectors including pBR322 pUC18, and pRC19c, which have become key tools of genetic engineering in the laboratory and beyond, such as in agriculture.”
Although the blá gene sequence of the synthetic plasmid vector (locus 3293-4153) is identical to that of the wild type plasmid, a substantial difference in the sequence upstream of the blá gene was apparent. Specifically, the transposase gene present in the wild-type Tn3 was lost and the pBR322 locus 1763-3146 was obtained from another plasmid, pMB1. These distinctive sequence features make it possible to distinguish the synthetic plasmid vector-sourced blá from the wild-type plasmid-sourced blá using sequence-based methods, such as PCR with primers designed to span the loci of 1773-3146 and 3293-4153. Furthermore, the pBR322 locus 2347-4353 was found to be shared with pUC19. Hence, in their study, synthetic plasmid vector-specific PCR and qPCR primers targeting the regions of 3086-3448 and 3243-3305 were used to survey the existence and distribution of vector-sourced ampicillin-resistant gene contamination in environmental microbes. And to their surprise, the blá gene from the synthetic plasmid vector was detected in samples taken from all six of the rivers.
Environmental samples are drawn from a complex and dynamic bacterial community and a large portion of the species cannot be cultured. Even in those that can be cultured, not all plasmids can be captured. Metagenomic technology, which involves transforming environmental genomic DNA into a lab strain, is a unique way of studying complex genetic samples from ecosystems without purifying the strains. It has recently been proven useful in analysing a diversity of environmental samples. This method has been adapted for their study. Plasmids were extracted directly from environmental microbes for transformation of lab E. coli, and a plasmid metagenomic library of 205 ampicillin resistance strains constructed, of which 27.3 % were positive for blá.
They concluded (p. 13454): “the potential hazards of environmental release of synthetic plasmid vectors and genetically modified products containing the vector components should be given more attention.”
The World Health Organisation has released its 2014 report on antimicrobial resistance, sounding appropriate alarm that the last resort antimicrobial has been breached, and exhorting the world’s nations rightly to avoid overuse and abuse of antibiotics [14]. However, it has omitted mentioning GMOs and the GM technologies altogether, which as the Chinese study has demonstrated, is a major source of antibiotic resistance affecting humans and livestock. The importance of genetic modification in spreading antibiotic resistance was predicted in our paper published in 1998 [1].
Molecular geneticists in Europe, USA, and elsewhere should now probe for the presence of GM nucleic acids including antibiotic resistance marker genes in the environment while halting the further releases of GM nucleic acids, both deliberate and ‘tolerated’ from ‘contained use’.
Article first published 09/06/14
Got something to say about this page? Comment
There are 5 comments on this article so far. Add your comment above.
Rory Short Comment left 10th June 2014 01:01:42
I personally am not surprised by these findings. It took nature 3.8 billion years to arrive at humankind and yet there are some among us who think that they know enough to safely meddle with nature. Their arrogance is off any measurable scale.
Ken Lys Comment left 11th June 2014 06:06:24
More proof that man's knowledge far exceeds his wisdom. Monsanto is hell bent on being first in this market capturing it all first then (maybe)sorting out the issue of health and safety along the way as it comes up. That means Monsanto makes all the profit, and we are to blindly take all the risk! Standard corporate philosophy - nothing and no one matters but the 90 day projection. BOYCOTT EVERYTHING GMO until they stop Monsanto's madness.
Susan Comment left 8th July 2014 16:04:33
Thank you for a brilliant report. I agree with the 2 comments before me but we must do more than comment, we must all begin to share these reports with everyone we know. They deserve the truth don't they? And we all need to share this information with our Representatives and let them know that they are on notice and will be held accountable for the impacts of genetically screwing up everything. We do not consent to being an experiment! Contact yours here:
✎ ☏ Find your US Representatives by Zip Code:
http://www.contactingthecongress.org/
If they don't listen, why don't we vote them out? Way past due I think...
###
Don Mau Comment left 12th October 2014 02:02:35
God help us all.
Agrobacterium , a rhizobiales order bacteria, and Bartonella , a rhizobiales order bacteria use HGT to give Bartonella upgrades of pathogenic virulence.
http://microbewiki.kenyon.edu/index.php/Bartonella_henselae
“Bartonella are the only bacteria able to produce angiogenic tumors in humans, very much like the Agrobacterium species that produce tumors in plants”
http://www.townsendletter.com/July2009/ed_lyme0709.html
"The knowledge base about both Bartonella testing and treatment borders on the disastrous. Bartonella is one of the most common infections in the world. Calling it a "coinfection" is nonsense; if anything, Lyme is the "coinfection." It is found in vast numbers of common vectors, including dust mites, fleas, flea feces, pet saliva, and ticks. Amazingly, it can turn off or lower antibodies to Lyme disease, Babesia, Ehrlichia, Anaplasma, and even itself. Bartonella floats in blood and also enters all blood vessel walls without causing a fatal fever, and indeed actually lowers fevers. It is the ultimate stealth infection. It turns off antibodies, fevers, and immune function defense chemicals as it damages organs in 20 to 60 ways."
In other words the "Lyme" disease epidemic is a direct result of Genetic engineering.
Curing "Lyme" is complex and a multifaceted approach is required.
My work on how to accomplish this;
www.lyme-morgellons.com
saptaparni roy chowdhury Comment left 4th July 2015 20:08:15
nice to read this article