Whole microbial genomes can be changed at will simultaneously and rapidly; with large implications for safety Dr. Mae-Wan Ho
One major way in which synthetic biology goes beyond conventional genetic engineering is in the manipulation of entire genomes, both in assembling it from pieces, as genome sequence data become available, and in modifying genes scattered over entire genomes. It must be emphasized that these techniques work as intended only in microbes where recombination between homologous (similar) nucleic acid sequences is the rule. In plants and animals, however, non-homologous recombination between dissimilar sequences predominates, and it is very difficult to target genes precisely. That is why genetic modification of crops and livestock is inherently uncontrollable and hazardous, as people like me have been stressing since it all began; and we have been proved right (see [1] GM is Dangerous and Futile, SiS 40). Synthetic biologists must proceed with caution in targeting plant and animal genomes, because the unpredictability and potential hazards are multiplied many times over.
The motivation for genome engineering, according to researchers Peter Carr at Massachusetts Institute of Technology, and George Church at Harvard Medical School, Cambridge, Massachusetts [2], is to understand through building, to produce medicines and biofuels with a genome optimized for the purpose, for example, to use microbes as biosensors, for bioremediation, to hunt and destroy cancer cells; to instruct our own cells to minimize the risk of septic shock; to produce organisms with fundamentally altered codon usage, which could prevent an engineered lab strain using acquired genes, and donating its engineered features to wild organisms. Or simply, the motivation is “build to creatively explore.”
Examples of genome engineering include Craig Venter Institute’s construction of the first microbial genome from commercially available cassettes [3]; the deletion of many large segments of E. coli genome to get rid of unstable DNA elements; the transfer of much of one Archean bacteria genome into eubacteria genome; decomposing the T7 bateriophage genome into many reconfigurable modules; making large numbers of targeted changes to a genome simultaneously (see below); and developing a purified translation system useful for in vitro prototyping of genetic functions without requiring moving genes into living cells. The manipulation of DNA segments >100 kbp in the first three examples relied heavily on in vivo recombination techniques.
One key technique that made it possible to manipulate entire genomes is the synthesis of DNA from scratch. Since the first synthetic gene was produced in the 1970s, the size of synthesized DNA increased exponentially until the early 2000s, when it experienced super-exponential growth with the construction of the entire 582 960 bp genome of Mycoplasma genitalium (see Figure 1a) [2]. Also since the early 2000s, the cost of sequencing and synthesis of DNA has dramatically decreased (see Figure 1b). It is possible, by 2009, to produce DNA at 100kbp/dollar, and sequence 1 Mbp/dollar (top two curves, Fig. 1b). But getting a novel dsDNA (double stranded DNA) gene construct to work by design is a nontrivial process, and that is lagging far behind (bottom curve, Fig. 1b).
“Genome engineering is in its infancy.” Carr and Church said [2]. The new techniques have enabled initial work to be done, but more sophisticated tools are needed at all stages: “design, DNA construction and manipulation, implementation and testing, and debugging.”
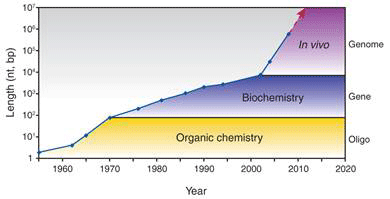
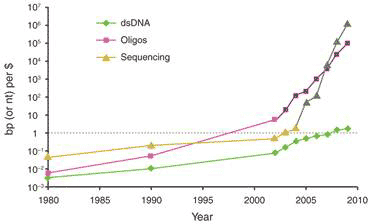
Figure 1 Growth in DNA technologies since genetic engineering began in the 1970s
The preferred procedure for genome manipulation is variation and selection instead of specific design, a kind of super-accelerated evolution. This is where mass genome engineering comes in, which has two big potential advantages: the preexistence of highly evolved modules in cells and microorganisms, and the possibility of introducing combinatorial and other genome-wide changes in the lab with high throughput techniques. One general category focuses on improving existing functions or selecting for new functions through directed evolution.
Computer-aided design (CAD) tools for high level design and simulation are crucial, as well as the detailed layout and sequences of oligonucleotides for multiplex assembly of genes or genomes. CAD specifies the complete combinations of genetic modifications for metabolic engineering and for sequence-based screening, where the number of changes to be made is too large, such as the genome-wide codon conversion in E. coli, where all TAG stop codons are to be converted to TAA (another stop codon). CAD tools are also needed to generate metabolic and signaling pathways, including processes not yet found in nature. A major goal for the future is to automate and integrate various aspects [2] “from protein design to compatibility of standards and intellectual property.”
DNA synthesis companies generally use fluid handling robots and moderately high density (96-. 384- or 1 536-well) reaction plates, which can produce megabase pairs of DNA per month. Another approach is microfluidic processing, which minimizes reagent and consumables and depend on parallel synthesis of shorter sequences that are assembled into longer ones.
Multiplex automated genome engineering (MAGE) is being developed for the large-scale programming and evolution of cells [3]. MAGE simultaneously targets many sites on the chromosome for modification in a single cell or across a population of cells, thus producing vast combinations of genomic diversity to select for the optimum combinations. The process is cyclical and scalable. The research team led by George Church constructed prototype devices that automate MAGE to facilitate rapid and continuous generation of mutations. MAGE was used to optimize the 1-deoxy-D-xylulose-5-phosphate (DXP) biosynthesis pathway in E coli to overproduce lycopene. Mediated by the bacteriophage l-Red ssDNA-binding protein b, gene replacement is achieved in the E. coli strain by directing ssDNA oligonucleotides to the lagging strand of the replication fork during DNA replication. The ssDNA were introduced by electroporation.
Twenty genes, documented to increase lycopene yield were targeted in an attempt to optimize translation by 90 base oligonucleotides containing degenerate ribosome binding site sequences flanked by homologous regions on each side. In addition, four genes from secondary pathways were targeted for inactivation by replacement sequences with stop codons to further improve flux through the DXP pathway. Thus, a total of 24 genes were optimized simultaneously for lycopene production.
As many as 15 billion genetic variants were generated. Screening was done by isolating colonies that produced intense red pigmentation (of lycopene) on agar plates. Sequencing of six variants revealed ribosomal binding site consensus Shine-Dalgarno sequence (which effectively recruits ribosome to start translation) in genes located at the beginning and end of the biosynthesis pathway; and gene knockouts from secondary pathways. The yield of lycopene was up to 9 mg/g dry cell weight.
In a later application using a general co-selection strategy with genetic markers within ~500kb of the targeted sites, the simultaneous modification of 80 sites in the E. coli genome was achieved [4].
MAGE combines targeted genome engineering with selection to rapidly create novel genomes with the required characteristics, and is the most effective genome modification process on the horizon. Such massive changes to genomes are bound to have unintended effects through gene, epigenetic and metabolic interactions, and it is vitally important that the new strains created are thoroughly characterized and risk assessed.
Although the process takes place presumably under contained use, how strict is the containment? Are transgenic wastes – containing tens of billions of new genomes created at any one time – discharged into the environment? These new genomes would offer numerous opportunities for horizontal gene transfer and recombination to create yet further new strains of bacteria and viruses, at least some of which may be serious pathogens. It is crucial to prevent this from happening.
Article first published 22/10/12
Got something to say about this page? Comment
There are 2 comments on this article so far. Add your comment above.
Warren Brodey M.D. Comment left 23rd October 2012 04:04:18
Have we enough wisdom to control the powerful tools we have created? Ask yourself and your family, your close friends and then your colleagues When the cost of an error is too high should I limit my creativity?
Is this a new question?
I believe it is.
Our power to destroy humanity has a new dimension. We can succeed.
Harold Titsonbeli sr Comment left 24th October 2012 05:05:07
Thank you for this,it makes connections possible to emerging diseases.We only find out what these mad scientists are doing after consideral time has past.Where is the power for an international ethics review?I am a very curious character,I always run in to unintended effects,some minor at first,however hindsight knowledge requires one to exist to review info.The benifits of this science is only to carry out depop.So it must be agreed upon by all partys' involved,not just those in the loop.My understanding of progress,wide diffinition,is that yields of modifications are based on best enviormental factors,not adversity.This comes to a bad end with gm s having a greater demand on food and water for optimum yeilds,so what happens in adverse conditions,meaning greatest time of need,drought?massive die off.This knowledge is at the second meta level,what about the next. Meta levels???? There is no impending doom to justify this effort ,other than prepare organisms to survive on ther planets ,or drastic ,maybe intended,changes to our biom