Increased longevity passed down generations of worms through epigenetic changes despite unaltered genomes Dr Eva Sirinathsinghji
New research finds that increased life-span can be inherited in a non-genetic manner. The study [1] by US researchers Anne Brunet and colleagues at Stanford University, Palo Alto, California, and Harvard University, Boston, Massachusetts, adds to the growing evidence of epigenetic inheritance that increasingly undermines the conventional idea that genes determine traits.
Epigenetic inheritance is usually described as the inheritance of a trait without inheriting any changes in the DNA sequence itself. These epigenetic changes can occur through various mechanisms including the chemical modification of DNA, RNA or proteins (commonly histone proteins that interact with, and determine the structural organisation of, DNA). More recently, other mechanisms of epigenetic changes are becoming widely recognised. Examples include RNA-editing, where RNA sequence bases are systematically altered, and alternative splicing of RNA to create different proteins, as well as RNA interference, which determines which RNA messages are cleaved or blocked from being translated into proteins (see [2] Epigenetic Inheritance “What Genes Remember”, SiS 41). Things are clearly a lot more complicated and dynamic than the old idea that DNA in the genome is fixed and determines all the traits of the organism in a series of linear instructions.
Numerous proteins have been implicated in controlling life-span. Among them are those that modify chromatin, a combination of DNA wrapped around histone proteins [3-5]. Deletions of certain genes that modify chromatin can lead to increased longevity.
The new work [1] shows that a ‘memory’ of those deletions can be passed on to descendents and increase their life-span without inheriting the actual mutation responsible for the increased longevity. This is the first evidence of epigenetic inheritance of a complex trait such as longevity.
Using the worm Caenorhabditis elegans as the model animal, the researchers deleted genes associated with a protein complex called COMPASS, which regulates a specific kind of histone protein modification, the trimethylation of lysine 4 of histone 3 (H3K4me3). In general, histone modifications serve as a chemical mark that influences the way DNA is packaged, and the fully methylated H3K4me3 is specifically associated with active, or ‘switched’ on genes. Altering the chromatin structure is an important determinant of gene expression, with compacted chromatin being a physical barrier to the transcriptional machinery. Deletion of the genes wdr-5, ash-2 and set-2, components of COMPASS led to reduced H3K4me3 levels, a change in gene activity, and a corresponding 20-30 % increase in the worms’ life-span.
Crucially, mating these longer-living mutant worms to normal (wild-type) worms gave rise to great-grand-offspring and even great-great-grand-offspring that no longer carried the gene deletion, but nevertheless still inherited the longer-living trait. Only after five generations following the creation of the knock-out worms (or 4 in the case of ash-2 mutations) did the worms reverted to the normal length life-span (see Figure 1).
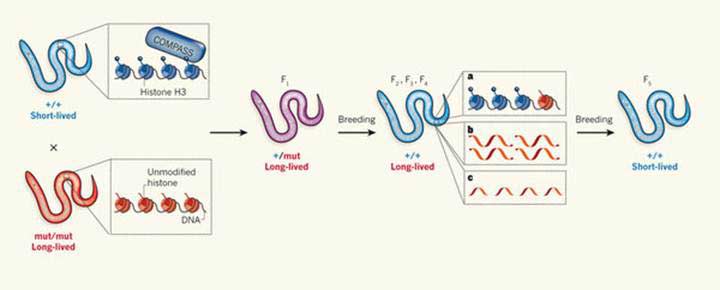
Figure 1 Memory of mutant gene lingers on even in complete absence of the mutant gene through four successive generations, adapted from Mango 2011 [6]
As can be seen, short-lived, wild-type (+/+) worms have two copies of functioning COMPASS complex gene that mediates how DNA is organised and how much genes are switched on. Mutant worms with two copies of the mutant gene (-/-), generated by the researchers, lack components of the COMPASS protein complex and live longer as a result. When wild-type worms are mated with mutant worms, their offspring (F1 generation) are heterozygous (+/-) for the mutation, carrying only one copy of the COMPASS components, but still live longer. When heterozygous F1 offspring are bred to wild-type worms, their offspring (F2 generation) that are completely wild-type for the mutation (+/+), still live longer despite no longer carrying the mutation. The same longer-living phenotype persisted in F3 and F4 generations. By the F5 generation however, the longer-living phenotype was lost and the worms reverted to the normal short-lived phenotype.
So, although the genetically wild-type worms were not carrying the mutation to make them live longer, they inherited a ‘memory’ of the mutation through epigenetic marking of histone proteins that was enough to increase their life-span. Previously, it was thought that most, if not all epigenetic changes would be wiped clean in the germ cells, thus preventing the inheritance of chemical modifications and acquired traits built up in one individual’s life-time. This finding, among others, suggests that complete re-programming of epigenetic modifications does not occur, with some modifications passing down the germ line to the offspring. From an evolutionary perspective, the ability to inherit learned or acquired traits from ancestors could be beneficial, increasing the chances of survival of the children who encounter similar life experiences to their parents.
Deletion of another gene rbr-2 associated with demethylation of lysine 4 of histone 3 abolished the effects of the mutations, suggesting that the results were not due to other, unrelated mutations that may occur in the experimental process.
In the nucleus of eukaryotic cells, DNA is organised and compacted in the form of chromatin that is wrapped around proteins called histones. Histone modification alters the structure of chromatin and how tightly the DNA is wrapped around the histones, thereby making genes either more or less accessible to the transcription machinery responsible for gene expression. These modifications can also deter the binding of transcriptional machinery to DNA and stop transcription altogether. Further, H3K4me3 is associated with transcriptionally ‘active’ genes and not ‘silent’ or switched off genes. When the researchers depleted the levels of H3K4me3 by deleting any of the genes responsible for adding the methyl groups, increased longevity was observed, possibly through changing the expression of genes that mediate life-span. Microarray experiments, which simultaneously measure gene expression of many or all genes to give a global gene expression profile, confirmed that these mutations did indeed lead to changes in gene expression on a genome-wide scale. Even more interesting was that the changes in gene expression observed in the mutant ancestors were similar to the genetically normal longer-living descendents. Some of the genes affected have previously been implicated in longevity, development and growth.
Still not understood is whether the H3K4me3 marks themselves or downstream events are the inherited cue for longevity. The study showed that the global reduced levels of H3K4me3 found in the mutant ancestors were not passed on. It is however possible that the chromatin changes at specific gene loci were not completely reset between generations. Alternatively, the RNA or proteins of downstream mis-expressed genes involved in longevity could also be passed on via the new oocytes to the next generation.
This study highlights the importance of environmental factors in the inheritance of complex traits. Our interaction with the world could have effects well beyond our life time.
Article first published 08/05/12
Got something to say about this page? Comment