Water’s Quantum Jazz
Other reports in this series
How water keeps ‘most everything in the cell dancing most of the time, and what’s the cell really like Dr. Mae-Wan Ho
We are now ready to venture inside the cell and see how everything, the rainbow ensemble, can do water’s quantum jazz together. We start by considering the interaction of water with ions and proteins.
The interaction of charged ions with water and proteins is at the heart of many signal transduction processes in the cell. Enzymes and cofactors are highly specific in their requirements for metal ions; while the addition of a phosphate group to proteins and metabolites – phosphorylation - are widely involved in activating enzyme pathways of biosynthesis and energy metabolism. What is the origin of these ion-specific effects? At a more fundamental level, why do different salts vary so much in solubility? And why do some salts precipitate proteins from solution more so than others?
Numerous studies have confirmed that small ions of high charge density are kosmotropes (order inducing) and bind water molecules strongly; while large ions of low charge density are chaotropes (disorder inducing) and bind water molecules weakly relative to the strength with which water molecules form hydrogen bonds with one another (see [1] Dancing with Ions, SiS 48). Kosmotropes tend to attract a solvation shell with more water molecules, while chaotropes have small solvation shells with less water molecules. But that is only half the story. The other half of the story begins with how ions interact with proteins.
Franz Hofmeister, a Czech scientist in the late 19th century, found that some salts helped egg white proteins to dissolve in water, while others caused the proteins to precipitate out, and there were those that had effects in between. He ranked the ions according to their ability to “salt-out” and “salt-in”, which resulted in the Hofmeister series. The Hofmeister series is also correlated with the ability of the ions to induce protein unfolding, coalescence of bubbles and many other phenomena, though there has never been a satisfactory explanation [2].
Kim Collins at University of Maryland Medical School, Baltimore, USA, may have found the answer [3, 4], and it is related to the ions’ affinity for water.
When pairs of oppositely charged ions have similar affinities for water, something special happens: they come out of their solvation shells, join up and neutralize each other. That’s because they can just as easily form intimate partners with each other as with water molecules; exchanging water molecules for the counter-ion does not cost anything in energetic terms. This ‘Law of Matching Water Affinities’ appears to explain why certain salts are less soluble than others, and why some salts precipitate proteins out of solution while others help them dissolve. The answer is that only neutral molecules precipitate (or crystallize) out of solution; neutral molecules have much lower solubility.
More specifically, according to Collins, a radius of 1.06 Ǻ separates small monovalent cations from large ones, and a radius of 1.78 Ǻ separates small monovalent anions from large ones. Small monovalent ions are strongly hydrated, while large monovalent ions are weakly hydrated (see Fig. 1). For example, LiF contains small monovalent ions that readily come out of their hydration shells to pair up as ‘contact ion pairs’, it has a solubility of only 0.1 M. In contrast, CsF has a large cation and a small anion, and do not pair up in solution; it has a solubility of 24.2 M.
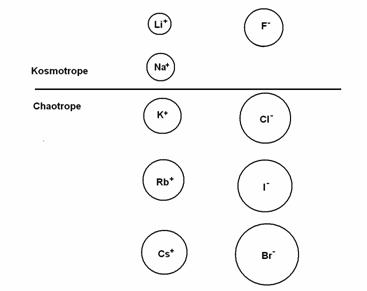
Figure 1 Monovalent kosmotropes & chaotropes ordered by size; the size of their hydrated ion is in inverse order
Proteins have strong negatively charged carboxylate groups (COO-) on their side chains that pair up well with kosmotrope cations, so Na+ salts out proteins, while a chaotrope such as Cs+ salts them in. Similarly, Ca2+ is well-matched to carboxylate in water affinity and will also salt out proteins.
Collin’s theory has received strong support from other scientists. Pavel Jungwirth and colleagues at the Czech Republic Academy of Sciences in Prague have quantified the higher affinity of Na+ over K+ for proteins by means of molecular dynamics simulations and conductivity measurements [5]. Both methods showed that sodium binds at least twice as strongly to the protein surface than potassium, and it is the same for all proteins studied. The carboxylate groups of aspartate and glutamate side chains played the major role. This same selective preference of the carboxylate groups for Na+ can be demonstrated in the isolated amino acids glutamic acid and aspartic acids, as well as in short peptides, and even in the simplest carboxylic acids, formate or acetate, as shown by molecular dynamics and quantum chemical calculations.
The research team led by Richard Saykally at US Department of Energy’s Lawrence Berkeley National Laboratory provided further support to Collin’s theory using the lab’s sophisticated x-ray probes of molecular bonds in samples ejected as microjets [6]. The selective binding of Na+ was demonstrated for acetate, though not for formate.
However, Berk Hess and Nico van der Vegt at the Max Planck Institute for Polymer Research in Mainz, Germany showed that Collin’s Law fails to describe the interaction of cations with carboxylate on the surface of many proteins [7]. Their molecular simulation results showed that the order of increasing binding affinity with carboxylate K+ < Na+ < Li+ is caused by a stronger preference for forming weak solvent-shared ion pairs – ion pairs that retain their solvation shells - rather than contact ion pairs that join up directly with each other. Contact pair interactions of these cations with protein surfaces, according to their simulations, are insignificant, indicating that the thermodynamic stability and interactions between proteins in alkali salt solutions is predominantly through hydration water molecules.
The most important explanation offered by Collin’s theory is why the ions present inside cells are so different from those outside, which has long puzzled biologists (see Table 1) [8]. Intracellular fluid has high concentrations of potassium and magnesium cations and phosphate and sulphate anions, and very low concentrations of sodium and chloride; the converse is true of extracellular fluid: low in potassium, magnesium, phosphate and sulphate, and high in sodium and chloride. While there appears to be not much difference between extracellular and intracellular calcium, most of the intracellular calcium is bound, with only 10-7M free Ca2+ most of the time, except for very transient, local increases associated with signal transduction.
|
Table 1 Ionic composition of intracellular and extracellular fluids (mM) | |||
| Ion | Intracellular | Extracellular | |
| Plasma | Interstitial | ||
| Cations | |||
| Potassium | 160 | 4 | 4 |
| Sodium | 10 | 142 | 145 |
| Calcium | 2* | 5 | 5 |
| Magnesium | 26 | 2 | 2 |
| Total cations | 198 | 153 | 156 |
Anions | |||
| Chloride | 3 | 101 | 114 |
| Bicarbonate | 10 | 27 | 31 |
| Phosphate | 100** | 2 | 2 |
| Sulphate | 20 | 1 | 1 |
| Organic acid | 0 | 6 | 7 |
| Protein | 65 | 16 | 1 |
| Total anions | 198 | 153 | 156 |
*Almost all intracellular calcium is bound, wiTR only 0.0001mM existing as free cation ** Free phosphate only 5 to 8 mM | |||
Apart from the inorganic ions, there are some 65 mM of proteins present in the cytoplasm, which are rich in carboxylate anions in their side chains. As Collins pointed out [4], the intracellular ions are optimised for mismatch in water affinities, so as to maintain high solubility of the proteins and other constituents of the cytoplasm at all times. That’s why quantum jazz of the rainbow ensemble is possible.
Increasingly, protein-folding disorders are being identified, including Alzheimer’s disease, Parkinson’s disease, transmissible spongiform encephalopathies (mad cow disease), Huntington’s disease, and type II diabetes, which have been linked to ligand binding and hydration [9]. In all likelihood, these diseases represent different failures in keeping almost all the molecular participants in cellular biochemistry dancing with water at any one time, so some of them end up salting out at inappropriate places.
In view of the high affinity of sodium ions for carboxylate, intracellular concentration of sodium is kept very low; and it is generally believed, by a Na+/K+ ATPase that pumps sodium out of the cell in exchange for potassium. RNA and DNA and membrane phospholipids are phosphate di-esters built upon the phosphate anion. Phosphate and carboxylate anions are the fundamental ions of the cell. Phosphate is also important in metabolism, where many small molecules are phosphorylated to keep them in the cell and to provide a “handle” for enzymes to bind onto. The nucleotide triphosphates (adenosine triphosphate, ATP and others), play an apparently critical and essential role in energy metabolism. Phosphate functions as a reversible marker in signal transduction, with phosphorylation activating or deactivates proteins. As mentioned earlier, Ca2+ is well matched to carboxylates, it is also well matched to phosphates
Ca2+ readily pairs with carboxylates and phosphates, forming insoluble complexes with important biological functions, but only in the right place and at the right time. Calcium carbonate in eggshells and oyster shells is highly insoluble, calcium oxalate (kidney stones) even more so, and calcium hydroxyphosphate in bones and teeth the most insoluble of all.
So, while the intracellular concentration of free phosphate is about 5-8 mM, the concentration of free calcium is 10-7M. To maintain this low concentration, it is pumped out of the cell or returned to intracellular stores.
The intracellular high concentration of potassium is well suited to keep phosphate in solution; the solubility of K2HPO4 is about 8.6M, while that of Na2HPO4 is 0.93M. K+ is also mismatched to carboxylates compared with Na+ as indicated by in vitro experiments mentioned earlier. Nevertheless, K+ may still form pairs with carboxylates under certain circumstances, as it is only a weak chaotrope (see Fig. 1) (see also later).
In contrast to K+, which has a water affinity lower than carboxylate and phosphate, that of Mg2+ is higher; and that also makes Mg2+ suitable as an intracellular ion. It predominantly relates to proteins and nucleic acids through shells of water molecules. ATP (adenosine triphosphate), the main energy intermediate in organisms, must be bound to a magnesium ion in order to be biologically active. What is called ATP is often actually Mg-ATP. Similarly, magnesium plays a role in the stability of all polyphosphate compounds in the cells, including those associated with DNA and RNA synthesis. Magnesium plays a major role in the folding and stabilization of RNA, both in a “diffuse” form without a specific binding site, and by binding tightly at discrete sites (not mediated by water).
The secrets of water are being unravelled to an unprecedented extent within the past few years. As described in reports of the present series, we now know that water has quantum properties even at room temperature, and may even be quantum coherent. We now know that water does play the lead role in the dynamics of proteins and DNA, and other macromolecules such as RNA that make the quantum jazz of life possible. There is good evidence that water does exist as a mixture of two states, high density water and low density water; and as Philippa Wiggins so brilliantly suggested, the spontaneous inter-conversion of these two forms of water may be what gives life its seemingly boundless ‘free energy’ (see [10] Two-States Water Explains All? SiS 32 and Wiggin’s recent report [11]).
But the big mystery remains. How did the cell manage to have just the right combination of ions inside, which is completely the opposite of what’s on the outside? How could the cell have evolved before it acquired all the complicated pumps and channels for all the ions?
Physiologist Gilbert Ling has stubbornly rejected the idea that it was all due to membrane pumps. Instead, he has maintained for more than half a century that the cytoplasm naturally excludes sodium and binds potassium. His association-induction hypothesis proposes that the major components of living protoplasm – water, proteins, and K+ - exist in a closely associated, high-energy state at ‘rest’ [12-14]. Purified proteins, he said, are not at all what proteins are like in the cell. Instead, within the cell, most if not all proteins are extended so that the peptide bonds along their polypeptide chains are free to interact with multiple layers of polarized water molecules and their carboxylic side chains similarly preferentially bind K+ over Na+. The other reason is the ubiquitous presence of ATP in the living cell.
Living protoplasm is full of ATP, which is bound to proteins at certain ‘cardinal sites’, according to Ling. These ATP-bound sites then induce changes in the electron density, ultimately of the entire polypeptide chain, including the side chains.
In the absence of ATP, proteins do tend to adopt secondary structures - alpha helix, or a beta pleated sheet - due to hydrogen bonding between peptide bonds in the same chain, which gives them a folded up conformation where they don’t interact maximally with water. However, when ATP is bound to the cardinal site, it tends to withdraw electrons away from the protein chain, thereby inducing the hydrogen bonds to open up, unfolding the chain and enabling it to interact with water.
For me, among the most persuasive evidence was the experiments he and his co-workers performed, showing that cells with cell membranes made permeable with detergents, or completely cut off at one end, nevertheless maintained their distinctive ionic compositions over long periods of time (see my review of his work [15] Strong Medicine for Cell Biology, SiS 24).
It is necessary to look at the cell again, not as a membrane bound entity containing various organelles suspended in an otherwise featureless cytoplasm still widely supposed to consist of proteins in aqueous solution. That view was strenuously refuted by Joseph Needham who cited extensively evidence in his book, Order and Life first published in 1936 [16] in support of meticulous molecular organisation in living protoplasm. Living protoplasm differs strikingly in many respects from the same proteins dissolved in water, including the ultraviolet absorption spectra (see Fig. 2). Needham also anticipated the discovery in my own lab by proposing that organisms are polyphasic liquid crystals.
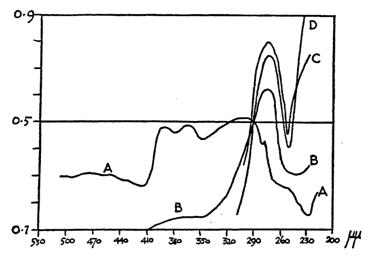
Figure 2 UV absorption for intake sea-urchin egg compared with solutions of proteins in water; A, intact egg; B, egg burst in water; C, crushed egg; D, solution of egg albumen [16]
Rickey Welch and James Clegg, lead champions of the view that the cytoplasm is organised; recently published an important review on the history of protoplasm [17]. They recall how Keith Porter used high voltage electron microscopy to reveal the ‘microtrabecular lattice’ (MTL) of the cytoplasm, which provides scaffolds for enzyme proteins and numerous micro-environments for specific enzyme reactions to take place independently of one another (Fig. 3).
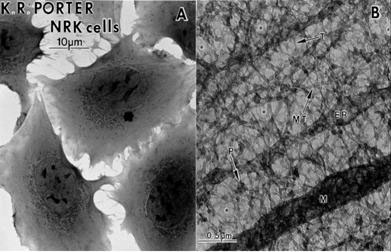
Figure 3 Newborn rat kidney cells A whole, B section showing microtrabecular lattice on high voltage electron microscopy [17]
Later, the MTL was rediscovered as the ubiquitous cytoskeleton of actin and other proteins, a dynamic structure that’s constantly breaking down and reforming as the cell changes shape, and transports materials to all parts of the cell. There is at least one journal entirely devoted to intracellular transport, full of photographs of cells criss-crossed by networks of neon-lit fibres that have been stained with fluorescent antibodies against various cytoskeletal proteins [18]
Martin Chaplin has proposed a scheme whereby ATP and K+ carboxylate binding are associated with actin polymerization and low density water with ice-like hydrogen bonds in a relatively static regime (or a resting, charged state), alternating with a more mobile high density water regime of actin depolymerisation, where hydrogen bonds are distorted or broken with less K+ carboxylate binding (see [19] The Importance of Cell Water, SiS 24) (Fig. 3).
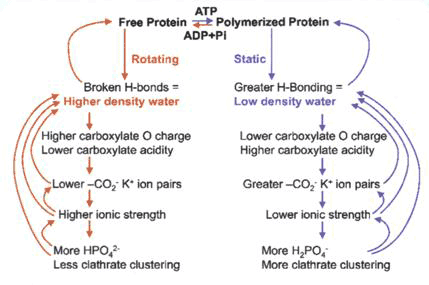
Figure 3 Change of state between high density and low density water as actin polymerises and depolymerises with associated changes in ion binding
These hypotheses can be tested by experiments. For example, as far as I know, no one has yet investigated whether proteins do behave differently in solutions of K+ and Mg-ATP. Or whether proteins enclosed in phospholipid micelles might show selectivity for K+ over Na+ due to a change in the state of water.
Most of all, we must bear in mind the hauntingly beautiful intimation of what the living cell is really like when Ludwig Edelmann at Saarland University in Hamburg, Germany, took extraordinary care in preserving its fully hydrated living configuration (Fig. 4, see also [20] What's the Cell Really Like? SiS 24).
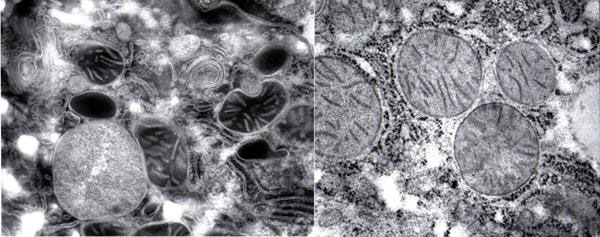
Figure 4 The fully hydrated cell (left) and the dehydrated cell (right) in conventional electron microscopy
Article first published 20/10/10
Comments are now closed for this article
There are 1 comments on this article.
Todd Millions Comment left 18th November 2010 08:08:58
dr-Mae-Wan Ho
Has any work being done on the two state water theory,and the non linear heat content of steam?
Though this was modelled in the 1800's(Rankine)-any structural reason for the complex heat content held by water in gas phase,in varing pressure and temptures has too my knowledge never being proposed.